Tutorial para gerar o mapa de distribuição de 𝑀𝑎𝑐𝑟𝑜𝑙𝑜𝑏𝑖𝑢𝑚 𝑎𝑟𝑎𝑐𝑎𝑒𝑛𝑠𝑒 Farroñay
By Ricardo de Oliveira Perdiz in mapa R Tutorial
September 1, 2019
Para executar o tutorial abaixo e reproduzir o mapa publicado em Farroñay et al. (2018), é necessário baixar os dados presentes na página
https://github.com/ricoperdiz/Tutorials/tree/master/R_map_2018_Farronayetal. Baixe todos os arquivos, com exceção dos arquivos .r e .ipynb (Jupyter Notebook).
Baixe o pdf na seção de
publicações.
01 Carrega os pacotes
Para manipular os dados e gerar o mapa desta postagem, fiz uso dos pacotes abaixo:
- broom (Robinson, Hayes, e Couch 2022);
- cowplot (Wilke 2020);
- dplyr (Wickham et al. 2021);
- ggsn (Santos Baquero 2019);
- GISTools (Brunsdon e Chen 2014);
- magrittr (Bache e Wickham 2020);
- measurements (Birk 2019);
- purrr (Henry e Wickham 2020);
- readr (Wickham, Hester, e Bryan 2021);
- rgdal (Bivand, Keitt, e Rowlingson 2021);
- sf (Pebesma 2021);
- stringr (Wickham 2019).
Tenha certeza de que todos os pacotes listados abaixo estão instalados.
Atenção especial deve ser dada ao pacote ggmap, que agora requer que seja instalado via GitHub. Veja mais detalhes
aqui. Para isso, instale a versão ggmap do GitHub rodando o comando abaixo:
# Rode o comando abaixo para instalar a versao do ggmap do GitHub
# Install ggmap package from GitHub version - this is needed in order to be able to download Google Maps nowadays
# if(!requireNamespace("remotes")) {
# install.packages("remotes")
# remotes::install_github("dkahle/ggmap")
# }
# ----------
## Atencao
# ----------
# ---------------------------------------------------------------
## Todos os dados necessarios para plotar o mapa desta publicacao estao disponiveis em:
# <https://github.com/ricoperdiz/Tutorials/tree/master/R_map_2018_Farronayetal>
## Script author: Ricardo Perdiz <ricardoperdiz@yahoo.com>
## Qualquer duvida ou erro, por favor deixe um alerta em <github.com/ricoperdiz/Tutorials/> ou escreva um email.
# ---------------------------------------------------------------
# lista de pacotes necessarios para rodar este tutorial
package_list <- c("dplyr", "stringr", "readr", "purrr", "magrittr", "broom", "rgdal", "measurements", "GISTools", "ggsn", "cowplot", "sf")
# Rode o comando abaixo para instalar todos os pacotes dentro do vetor package_list
# install.packages(package_list, dependencies = TRUE)
# Carregue os pacotes
libs <- c(package_list, "ggmap")
sapply(libs, library, character.only = TRUE, logical.return = TRUE, quietly = TRUE, warn.conflicts = FALSE)
## Warning: package 'broom' was built under R version 4.1.2
## rgdal: version: 1.5-23, (SVN revision 1121)
## Geospatial Data Abstraction Library extensions to R successfully loaded
## Loaded GDAL runtime: GDAL 3.2.1, released 2020/12/29
## Path to GDAL shared files: /Library/Frameworks/R.framework/Versions/4.1/Resources/library/rgdal/gdal
## GDAL binary built with GEOS: TRUE
## Loaded PROJ runtime: Rel. 7.2.1, January 1st, 2021, [PJ_VERSION: 721]
## Path to PROJ shared files: /Library/Frameworks/R.framework/Versions/4.1/Resources/library/rgdal/proj
## PROJ CDN enabled: FALSE
## Linking to sp version:1.4-5
## To mute warnings of possible GDAL/OSR exportToProj4() degradation,
## use options("rgdal_show_exportToProj4_warnings"="none") before loading rgdal.
## Overwritten PROJ_LIB was /Library/Frameworks/R.framework/Versions/4.1/Resources/library/rgdal/proj
## Checking rgeos availability: TRUE
## Please note that 'maptools' will be retired by the end of 2023,
## plan transition at your earliest convenience;
## some functionality will be moved to 'sp'.
## Warning: package 'MASS' was built under R version 4.1.2
##
## Attaching package: 'MASS'
## The following object is masked from 'package:dplyr':
##
## select
## rgeos version: 0.5-9, (SVN revision 684)
## GEOS runtime version: 3.8.1-CAPI-1.13.3
## Please note that rgeos will be retired by the end of 2023,
## plan transition to sf functions using GEOS at your earliest convenience.
## Linking to sp version: 1.4-6
## Polygon checking: TRUE
## Linking to GEOS 3.8.1, GDAL 3.2.1, PROJ 7.2.1
## Google's Terms of Service: https://cloud.google.com/maps-platform/terms/.
## Please cite ggmap if you use it! See citation("ggmap") for details.
## dplyr stringr readr purrr magrittr broom
## TRUE TRUE TRUE TRUE TRUE TRUE
## rgdal measurements GISTools ggsn cowplot sf
## TRUE TRUE TRUE TRUE TRUE TRUE
## ggmap
## TRUE
# If any of the messages below points FALSE, it means that you do not have that package installed. So, please install it before proceeding with the tutorial.
# if you do not remember how to install a package, use the command below
# install.packages("PackageName", dependencies = TRUE)
Os pacotes blogdown (Xie, Dervieux, e Presmanes Hill 2021), rmarkdown (Xie, Allaire, e Grolemund 2018), e knitr (Xie 2021) foram utilizados tanto para gerar a estrutura deste sítio web quanto para gerar os arquivos .html que vocês estão lendo neste momento.
02 Lê os dados
Dados para este tutorial se dividem entre dados dos espécimes e dados SIG. Dados de espécimes contêm coordenadas, enquanto dados SIG contêm todos os shapefiles necessários para os mapas.
Dados de espécimes
# Macrolobium longipes
nybg <- read_delim("https://raw.githubusercontent.com/ricoperdiz/Tutorials/master/R_map_2018_Farronayetal/nybg.csv", delim = ",")
## Warning: One or more parsing issues, see `problems()` for details
## Rows: 4 Columns: 33
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (20): AdmGUIDValue_tab, DarInstitutionCode, DarDatasetName, DarBasisOfRe...
## dbl (6): DarCatalogNumber, DarCollector, DarYearCollected, DarDecimalLatitu...
## lgl (7): DarMinimumElevation, DarMaximumElevation, DarGeorefMethod, DarSubs...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
splink <- read_delim("https://raw.githubusercontent.com/ricoperdiz/Tutorials/master/R_map_2018_Farronayetal/splink.csv", delim = ";")
## Rows: 4 Columns: 76
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ";"
## chr (22): institutioncode, kingdom, descrindividuo, aux_nomecompunidgeo_inv...
## dbl (9): codcolbot, codespecime, numtombo, numcoleta, diacoleta, mescoleta...
## lgl (43): basisofrecord, phylum, class, ordem, fieldnumber, julianday, time...
## date (2): datelastmodified, dataultalter
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
inpa <- read_delim("https://raw.githubusercontent.com/ricoperdiz/Tutorials/master/R_map_2018_Farronayetal/inpa.csv", delim = ",")
## Rows: 2 Columns: 5
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (4): COORD_, HERBARIA, COLETOR, ESPECIE
## dbl (1): NUMERO_COL
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
# Macrolobium aracaense
araca <- read_delim("https://raw.githubusercontent.com/ricoperdiz/Tutorials/master/R_map_2018_Farronayetal/Maracaense.csv", delim = "\t")
## Rows: 2 Columns: 17
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: "\t"
## chr (10): COLETOR, ADDCOLL, COUNTRY, MINORAREA, MAJORAREA, GAZETTER, COORDEN...
## dbl (6): NUMERO, COLDD, COLM, COLY, LATITUDE, LONGITUDE
## lgl (1): SP1
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Dados SIG
# South and Central America shape
area_mapa <- rgdal::readOGR(dsn = "SAm_CAm_shape.shp")
## OGR data source with driver: ESRI Shapefile
## Source: "/Users/ricoperdiz/Documents/PROFISSIONAL/Sites/ricardoperdiz/content/blog/2020_03_29_R-map-Farronayetal-2018/SAm_CAm_shape.shp", layer: "SAm_CAm_shape"
## with 37 features
## It has 65 fields
# Brazilian states shapefile
estados <- st_read("BRASIL.shp")
## Reading layer `BRASIL' from data source
## `/Users/ricoperdiz/Documents/PROFISSIONAL/Sites/ricardoperdiz/content/blog/2020_03_29_R-map-Farronayetal-2018/BRASIL.shp'
## using driver `ESRI Shapefile'
## Simple feature collection with 27 features and 3 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: -73.83943 ymin: -33.77086 xmax: -34.8581 ymax: 5.38289
## CRS: NA
# Filtra apenas o estado do Amazonas dentro do objeto `estados`
br_amazonas <- estados %>% filter(ESTADO == "Amazonas")
br_amazonas
## Simple feature collection with 1 feature and 3 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: -73.68224 ymin: -9.847688 xmax: -55.99345 ymax: 2.198446
## CRS: NA
## UF ESTADO REGIAO geometry
## 1 AM Amazonas NO MULTIPOLYGON (((-63.47887 2...
br_amazonas_tidy <- st_geometry(br_amazonas)
br_amazonas_tidy
## Geometry set for 1 feature
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: -73.68224 ymin: -9.847688 xmax: -55.99345 ymax: 2.198446
## CRS: NA
## MULTIPOLYGON (((-63.47887 2.198446, -63.43955 2...
area_mapa_tidy <- tidy(area_mapa)
## Regions defined for each Polygons
area_mapa_tidy
## # A tibble: 61,879 × 7
## long lat order hole piece group id
## <dbl> <dbl> <int> <lgl> <fct> <fct> <chr>
## 1 -70.0 12.6 1 FALSE 1 0.1 0
## 2 -69.9 12.5 2 FALSE 1 0.1 0
## 3 -69.9 12.5 3 FALSE 1 0.1 0
## 4 -69.9 12.5 4 FALSE 1 0.1 0
## 5 -69.9 12.5 5 FALSE 1 0.1 0
## 6 -69.9 12.4 6 FALSE 1 0.1 0
## 7 -69.9 12.4 7 FALSE 1 0.1 0
## 8 -69.9 12.4 8 FALSE 1 0.1 0
## 9 -69.9 12.4 9 FALSE 1 0.1 0
## 10 -69.9 12.4 10 FALSE 1 0.1 0
## # … with 61,869 more rows
03 Limpa os dados
Primeiro, limparemos os dados da espécie Macrolobium longipes R.S. Cowan.
# Macrolobium longipes
# first data from inpa
inpa_modif <- inpa %>%
mutate(
lat = str_split(COORD_, "N") %>%
sapply("[[", 1) %>%
gsub("°", " ", .) %>%
gsub("’", " ", .) %>%
paste0(., " 0") %>%
gsub(" | ", " ", .) %>%
str_trim("both") %>%
measurements::conv_unit(., from = "deg_min_sec", to = "dec_deg"),
long = str_split(COORD_, "N") %>%
sapply("[[", 2) %>%
gsub("°", " ", .) %>%
gsub("’", " ", .) %>%
gsub("W", "0", .) %>%
str_trim("both") %>%
gsub(" ", " ", .) %>%
measurements::conv_unit(., from = "deg_min_sec", to = "dec_deg")
) %>%
dplyr::select(-COORD_)
inpa_modif
## # A tibble: 2 × 6
## HERBARIA COLETOR NUMERO_COL ESPECIE lat long
## <chr> <chr> <dbl> <chr> <chr> <chr>
## 1 INPA Huber 5849 Macrolobium longipes 1.58333333333333 66.55
## 2 INPA Rodrigues 10875 Macrolobium longipes 0.333333333333333 67.31666…
inpa_modif %>% glimpse()
## Rows: 2
## Columns: 6
## $ HERBARIA <chr> "INPA", "INPA"
## $ COLETOR <chr> "Huber", "Rodrigues"
## $ NUMERO_COL <dbl> 5849, 10875
## $ ESPECIE <chr> "Macrolobium longipes", "Macrolobium longipes"
## $ lat <chr> "1.58333333333333", "0.333333333333333"
## $ long <chr> "66.55", "67.3166666666667"
names(inpa_modif) <- tolower(names(inpa_modif)) # turn all column names as lower case
names(inpa_modif)
## [1] "herbaria" "coletor" "numero_col" "especie" "lat"
## [6] "long"
# then we get speciesLink data and select only columns we want to keep
longipes <- splink %>%
dplyr::select(
herbaria = institutioncode,
coletor, numero_col = numcoleta,
especie = taxoncompleto,
pais, estado = estado_prov,
local = descrlocal
) %>%
# then join them with data from INPA
full_join(inpa_modif, .) %>%
dplyr::select(-estado) %>%
# turn lat and long columns as numeric
mutate_at(
.vars = c("lat", "long"),
.funs = as.numeric
)
## Joining, by = c("herbaria", "coletor", "numero_col", "especie")
longipes
## # A tibble: 6 × 8
## herbaria coletor numero_col especie lat long pais local
## <chr> <chr> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 INPA Huber 5849 Macrolobium… 1.58 66.6 <NA> <NA>
## 2 INPA Rodrigues 10875 Macrolobium… 0.333 67.3 <NA> <NA>
## 3 JBRJ Maguire, B. 37590 Macrolobium… NA NA Venez… "Rio Siapa, …
## 4 JBRJ Maguire, B. 37572 Macrolobium… NA NA Venez… "Rio Siapa, …
## 5 JBRJ Maguire, B. 37643 Macrolobium… NA NA Venez… "Rio Siapa, …
## 6 JBRJ Maguire, B. 37590 Macrolobium… NA NA Venez… "Rio Siapa, …
longipes %>% glimpse()
## Rows: 6
## Columns: 8
## $ herbaria <chr> "INPA", "INPA", "JBRJ", "JBRJ", "JBRJ", "JBRJ"
## $ coletor <chr> "Huber", "Rodrigues", "Maguire, B.", "Maguire, B.", "Maguir…
## $ numero_col <dbl> 5849, 10875, 37590, 37572, 37643, 37590
## $ especie <chr> "Macrolobium longipes", "Macrolobium longipes", "Macrolobiu…
## $ lat <dbl> 1.5833333, 0.3333333, NA, NA, NA, NA
## $ long <dbl> 66.55000, 67.31667, NA, NA, NA, NA
## $ pais <chr> NA, NA, "Venezuela", "Venezuela", "Venezuela", "Venezuela"
## $ local <chr> NA, NA, "Rio Siapa, Casiquiare, Territorio Amazonas. ", "Ri…
# at last, we get nybg data and select and rename columns that we want to keep
nybg_modif <- nybg %>%
dplyr::select(
herbaria = DarInstitutionCode,
coletor = DarCollectorNumber,
numero_col = DarCollector,
local = DarLocality,
lat = DarDecimalLatitude,
long = DarDecimalLongitude,
pais = DarCountry
)
nybg_modif
## # A tibble: 4 × 7
## herbaria coletor numero_col local lat long pais
## <chr> <chr> <dbl> <chr> <dbl> <dbl> <chr>
## 1 NY H. C. de Lima 3279 Alto Rio Negro, Rio U… -0.130 -67.1 Brazil
## 2 NY B. Maguire 37590 On right bank of Río … NA NA Venez…
## 3 NY B. Maguire 37590 On right bank of Río … NA NA Venez…
## 4 NY W. A. Rodrigues 10875 Alto Rio Negro. Igara… 0.333 -67.3 Brazil
Sempre é bom olhar nossos dados, checar o tipo de variável para cada coluna, quantas observações e quantas variáveis temos em nossos dados.
Podemos fazer isso utilizando a função str() do pacote base do R, ou a função glimpse do pacote dplyr.
Aqui usarei a função glimpse no conjunto de dados longipes.
glimpse(longipes)
## Rows: 6
## Columns: 8
## $ herbaria <chr> "INPA", "INPA", "JBRJ", "JBRJ", "JBRJ", "JBRJ"
## $ coletor <chr> "Huber", "Rodrigues", "Maguire, B.", "Maguire, B.", "Maguir…
## $ numero_col <dbl> 5849, 10875, 37590, 37572, 37643, 37590
## $ especie <chr> "Macrolobium longipes", "Macrolobium longipes", "Macrolobiu…
## $ lat <dbl> 1.5833333, 0.3333333, NA, NA, NA, NA
## $ long <dbl> 66.55000, 67.31667, NA, NA, NA, NA
## $ pais <chr> NA, NA, "Venezuela", "Venezuela", "Venezuela", "Venezuela"
## $ local <chr> NA, NA, "Rio Siapa, Casiquiare, Territorio Amazonas. ", "Ri…
glimpse(nybg_modif)
## Rows: 4
## Columns: 7
## $ herbaria <chr> "NY", "NY", "NY", "NY"
## $ coletor <chr> "H. C. de Lima", "B. Maguire", "B. Maguire", "W. A. Rodrigu…
## $ numero_col <dbl> 3279, 37590, 37590, 10875
## $ local <chr> "Alto Rio Negro, Rio Uaupés, Igarapé Tibuarí, acima do Seri…
## $ lat <dbl> -0.130278, NA, NA, 0.333333
## $ long <dbl> -67.0892, NA, NA, -67.3167
## $ pais <chr> "Brazil", "Venezuela", "Venezuela", "Brazil"
longipes <-
longipes %>%
full_join(., nybg_modif)
## Joining, by = c("herbaria", "coletor", "numero_col", "lat", "long", "pais", "local")
longipes # now with data from INPA and NY herbaria
## # A tibble: 10 × 8
## herbaria coletor numero_col especie lat long pais local
## <chr> <chr> <dbl> <chr> <dbl> <dbl> <chr> <chr>
## 1 INPA Huber 5849 Macrolob… 1.58 66.6 <NA> <NA>
## 2 INPA Rodrigues 10875 Macrolob… 0.333 67.3 <NA> <NA>
## 3 JBRJ Maguire, B. 37590 Macrolob… NA NA Venez… "Rio Siapa…
## 4 JBRJ Maguire, B. 37572 Macrolob… NA NA Venez… "Rio Siapa…
## 5 JBRJ Maguire, B. 37643 Macrolob… NA NA Venez… "Rio Siapa…
## 6 JBRJ Maguire, B. 37590 Macrolob… NA NA Venez… "Rio Siapa…
## 7 NY H. C. de Lima 3279 <NA> -0.130 -67.1 Brazil "Alto Rio …
## 8 NY B. Maguire 37590 <NA> NA NA Venez… "On right …
## 9 NY B. Maguire 37590 <NA> NA NA Venez… "On right …
## 10 NY W. A. Rodrigues 10875 <NA> 0.333 -67.3 Brazil "Alto Rio …
longipes <-
longipes %>%
filter(!is.na(lat), coletor != "Rodrigues") %>%
# add species name in a separate column
mutate(
especie = "M. longipes"
)
glimpse(longipes)
## Rows: 3
## Columns: 8
## $ herbaria <chr> "INPA", "NY", "NY"
## $ coletor <chr> "Huber", "H. C. de Lima", "W. A. Rodrigues"
## $ numero_col <dbl> 5849, 3279, 10875
## $ especie <chr> "M. longipes", "M. longipes", "M. longipes"
## $ lat <dbl> 1.583333, -0.130278, 0.333333
## $ long <dbl> 66.5500, -67.0892, -67.3167
## $ pais <chr> NA, "Brazil", "Brazil"
## $ local <chr> NA, "Alto Rio Negro, Rio Uaupés, Igarapé Tibuarí, acima do …
Agora limparemos os dados referentes à espécie nova descrita em Farroñay et al. (2018), chamada Macrolobium aracaense Farroñay.
# Macrolobium aracaense
## Clean data
# turn column names to lower case
names(araca) <- tolower(names(araca))
araca_md <- araca %>%
mutate(
# clean coordinate columns to extract and convert degrees min sec to decimal degrees
lat = str_split(coordenadas, "N") %>%
sapply("[[", 1) %>%
gsub("°", " ", .) %>%
gsub("\'|\'\'|\"", " ", .) %>%
gsub(" | ", " ", .) %>%
str_trim("both") %>%
measurements::conv_unit(., from = "deg_min_sec", to = "dec_deg"),
# do the same done for long data
long = str_split(coordenadas, "N") %>%
sapply("[[", 2) %>%
gsub("W", "", .) %>%
gsub("°", " ", .) %>%
gsub("\'|\'\'|\"", " ", .) %>%
gsub(" | ", " ", .) %>%
str_trim("both") %>%
measurements::conv_unit(., from = "deg_min_sec", to = "dec_deg"),
especie = "M. aracaense"
) %>%
dplyr::select(coletor,
numero_col = numero,
pais = country,
lat, long, estado = majorarea, herbaria, especie
) %>%
# turn lat and long columns as numeric
mutate_at(
.vars = c("lat", "long"),
.funs = as.numeric
)
glimpse(araca_md)
## Rows: 2
## Columns: 8
## $ coletor <chr> "Vicentini, A.", "Farroñay, F."
## $ numero_col <dbl> 1886, 178
## $ pais <chr> "Brasil", "Brasil"
## $ lat <dbl> 0.9483333, 0.8633333
## $ long <dbl> 63.39167, 63.33167
## $ estado <chr> "Amazonas", "Amazonas"
## $ herbaria <chr> "INPA", "INPA"
## $ especie <chr> "M. aracaense", "M. aracaense"
# join data for Macrolobium longipes and M. aracaense
dados <-
full_join(longipes, araca_md) %>%
dplyr::select(herbaria, coletor, numero_col, especie, lat, long)
## Joining, by = c("herbaria", "coletor", "numero_col", "especie", "lat", "long", "pais")
glimpse(dados)
## Rows: 5
## Columns: 6
## $ herbaria <chr> "INPA", "NY", "NY", "INPA", "INPA"
## $ coletor <chr> "Huber", "H. C. de Lima", "W. A. Rodrigues", "Vicentini, A.…
## $ numero_col <dbl> 5849, 3279, 10875, 1886, 178
## $ especie <chr> "M. longipes", "M. longipes", "M. longipes", "M. aracaense"…
## $ lat <dbl> 1.5833333, -0.1302780, 0.3333330, 0.9483333, 0.8633333
## $ long <dbl> 66.55000, -67.08920, -67.31670, 63.39167, 63.33167
dados$long <- ifelse(dados$long > 0, dados$long * -1, dados$long)
longipes <- dados %>%
filter(especie == "M. longipes")
aracaense <- dados %>%
filter(especie == "M. aracaense")
Agora temos dois objetos contendo os dados necessários para cada espécie, Macrolobium longipes e M. aracaense.
longipes
## # A tibble: 3 × 6
## herbaria coletor numero_col especie lat long
## <chr> <chr> <dbl> <chr> <dbl> <dbl>
## 1 INPA Huber 5849 M. longipes 1.58 -66.6
## 2 NY H. C. de Lima 3279 M. longipes -0.130 -67.1
## 3 NY W. A. Rodrigues 10875 M. longipes 0.333 -67.3
aracaense
## # A tibble: 2 × 6
## herbaria coletor numero_col especie lat long
## <chr> <chr> <dbl> <chr> <dbl> <dbl>
## 1 INPA Vicentini, A. 1886 M. aracaense 0.948 -63.4
## 2 INPA Farroñay, F. 178 M. aracaense 0.863 -63.3
04 Cria variáveis para o mapa
# longitude range - main map
x1 <- st_bbox(br_amazonas)[c(1, 3)]
x1
## xmin xmax
## -73.68224 -55.99345
y1 <- st_bbox(br_amazonas)[c(2, 4)]
y1
## ymin ymax
## -9.847688 2.198446
# longitude and latitude range - for ggmap
lat_long <- dados
y_geral <- range(lat_long$lat) + c(-0.01, 0.01)
y_geral
## [1] -0.140278 1.593333
x_geral <- range(lat_long$long) + c(-0.01, 0.01)
x_geral
## [1] -67.32670 -63.32167
05 Baixa o mapa via pacote R ggmap
Recentemente, houve uma mudança na maneira de baixar mapas Google através do pacote ggmap. Agora é necessária a obtenção de uma chave Google e utiizá-la ao rodar os comandos abaixo com a função mostrada abaixo. Após ese passo, o usuário terá a permissão de baixar os mapas Google usando a função ggmap::get_map.
Para mais informações, visite este sítio web.
# Place your google KEY here - without it, you WILL NOT be able to download a google map
# register_google(key = 'PlaceYourKeyRightHERE!')
# local_mp <- c(lon = sum(x_geral)/2, lat = sum(y_geral)/2)
# local_mp
# mapa <- get_map(location = local_mp,
# source = "google",
# maptype = "terrain", crop = FALSE,
# zoom = 7,
# color = 'bw',
# language = 'en-EN')
Para checar se funcionou, tente plotar o objeto mapa:
mapa
Se você não obteve sucesso em baixar o mapa, utilize o arquivo .RDS abaixo para poder prosseguir com o tutorial. Ele contem exatamente o mesmo mapa baixado do Google utilizado na publicação, e se encontra disponível na pasta https://github.com/ricoperdiz/Tutorials/tree/master/R_map_2018_Farronayetal.
mapa <- readRDS("Farronayetal2017_ggmap.RDS")
ggmap(mapa, extent = "panel")
06 Plotam-se os mapas
# Main map - species distribution
x_escala <- -65.1
araca_map1 <-
ggmap(mapa, extent = "panel") +
xlab("Longitude (WGS84)") +
ylab("Latitude")
# plot the map with X and Y Label on it
# take a first look to see if everything is all right
araca_map1
# Main map
araca_map <-
araca_map1 +
geom_point(aes(x = long, y = lat, shape = especie), data = dados, alpha = .8, color = "black", size = 4) +
# add scale bar on topleft
scalebar(
x.min = attr(mapa, "bb")[[2]],
y.min = attr(mapa, "bb")[[1]],
x.max = attr(mapa, "bb")[[4]],
y.max = attr(mapa, "bb")[[3]],
dist = 100, anchor = c(x = -66, y = -2.2),
model = "WGS84",
location = "topleft",
st.size = 4,
st.dist = 0.02,
transform = TRUE,
dist_unit = "km"
) +
# add North arrow
geom_segment(
arrow = arrow(length = unit(4, "mm"), type = "closed", angle = 40),
aes(x = x_escala, xend = x_escala, y = -1.7, yend = -1.3), colour = "black", size = 2
) +
annotate(
x = x_escala, y = -1.85, label = "N", color = "black", geom = "text", size = 6,
fontface = "bold"
) +
# change ggplot2 theme to Black and White
theme_bw() +
# change few details:
# color contour of panel border, legend position, plot margin etc
theme(
panel.border = element_rect(colour = "black", fill = NA, size = 1),
legend.title = element_blank(), legend.text = element_text(size = 12, face = "bold.italic"),
legend.position = "bottom",
plot.margin = unit(x = c(0.1, 0.1, 0, 0.1), units = "in")
)
araca_map
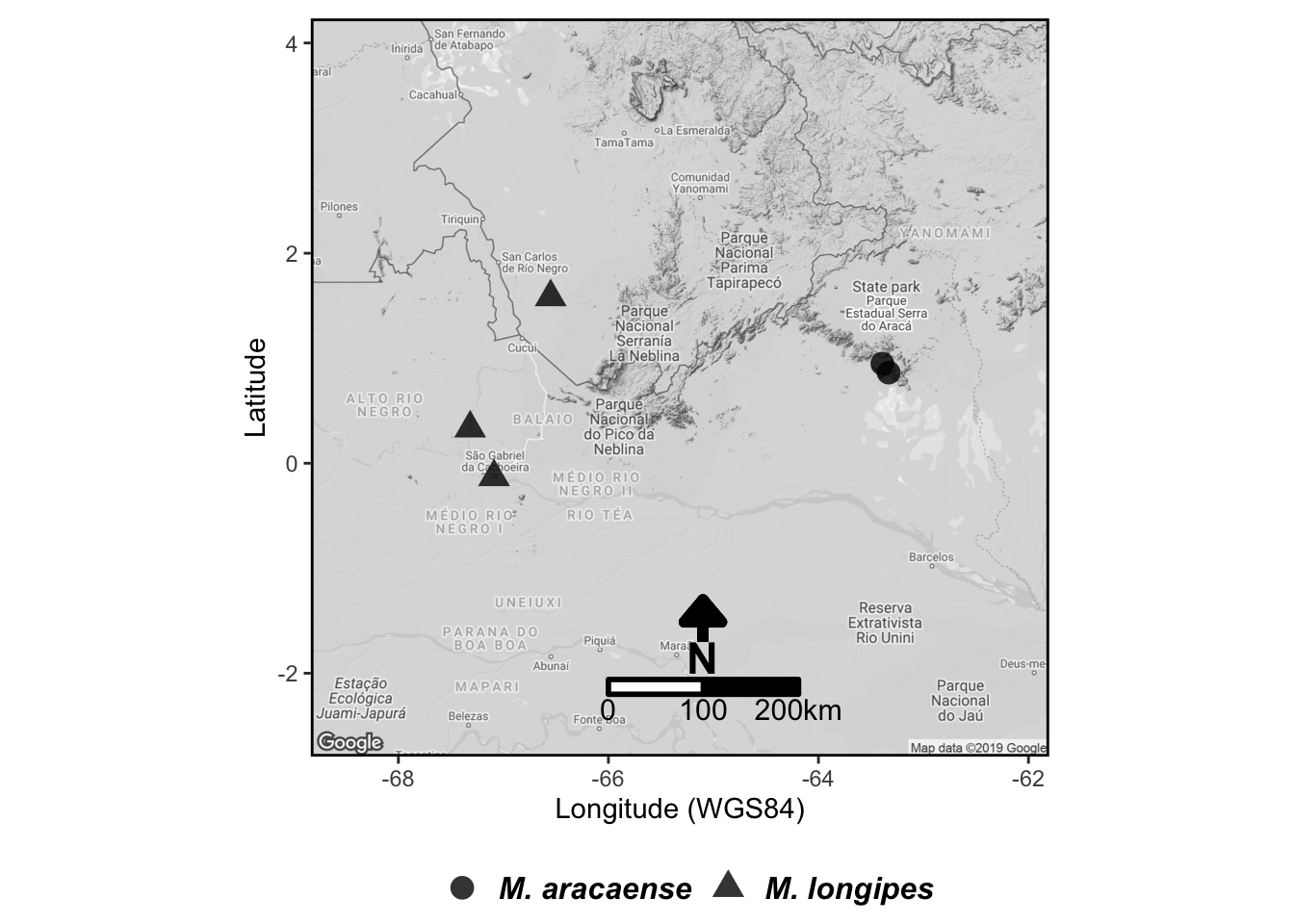
# plot the overview map
overviewmap1 <-
ggplot() +
geom_polygon(
data = as_tibble(st_coordinates(br_amazonas_tidy)),
aes(x = X, y = Y, group = L1),
color = "gray70", fill = "gray70", linetype = 3
)
overviewmap1
overviewmap2 <- overviewmap1 +
geom_polygon(
data = area_mapa_tidy,
aes(x = long, y = lat, group = group),
color = "black", fill = NA
) +
geom_point(
data = subset(dados, especie %in% "M. aracaense"),
aes(x = long, y = lat), size = 2,
pch = 19, col = "black"
)
overviewmap2
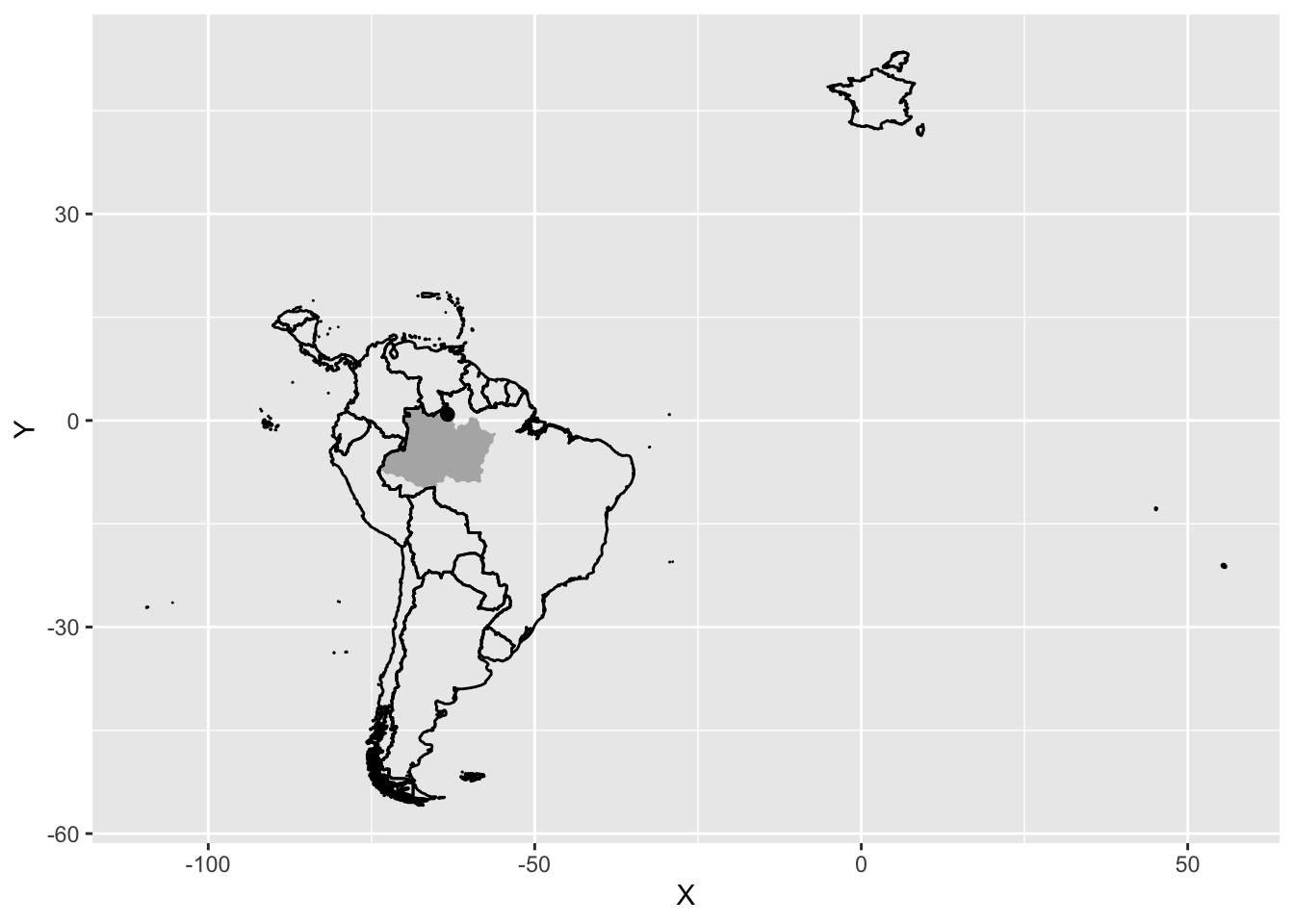
overviewmap3 <- overviewmap2 +
coord_equal() +
coord_cartesian(
xlim = x1 - c(2, -2), ylim = y1 - c(2, -2)
)
## Coordinate system already present. Adding new coordinate system, which will replace the existing one.
overviewmap3
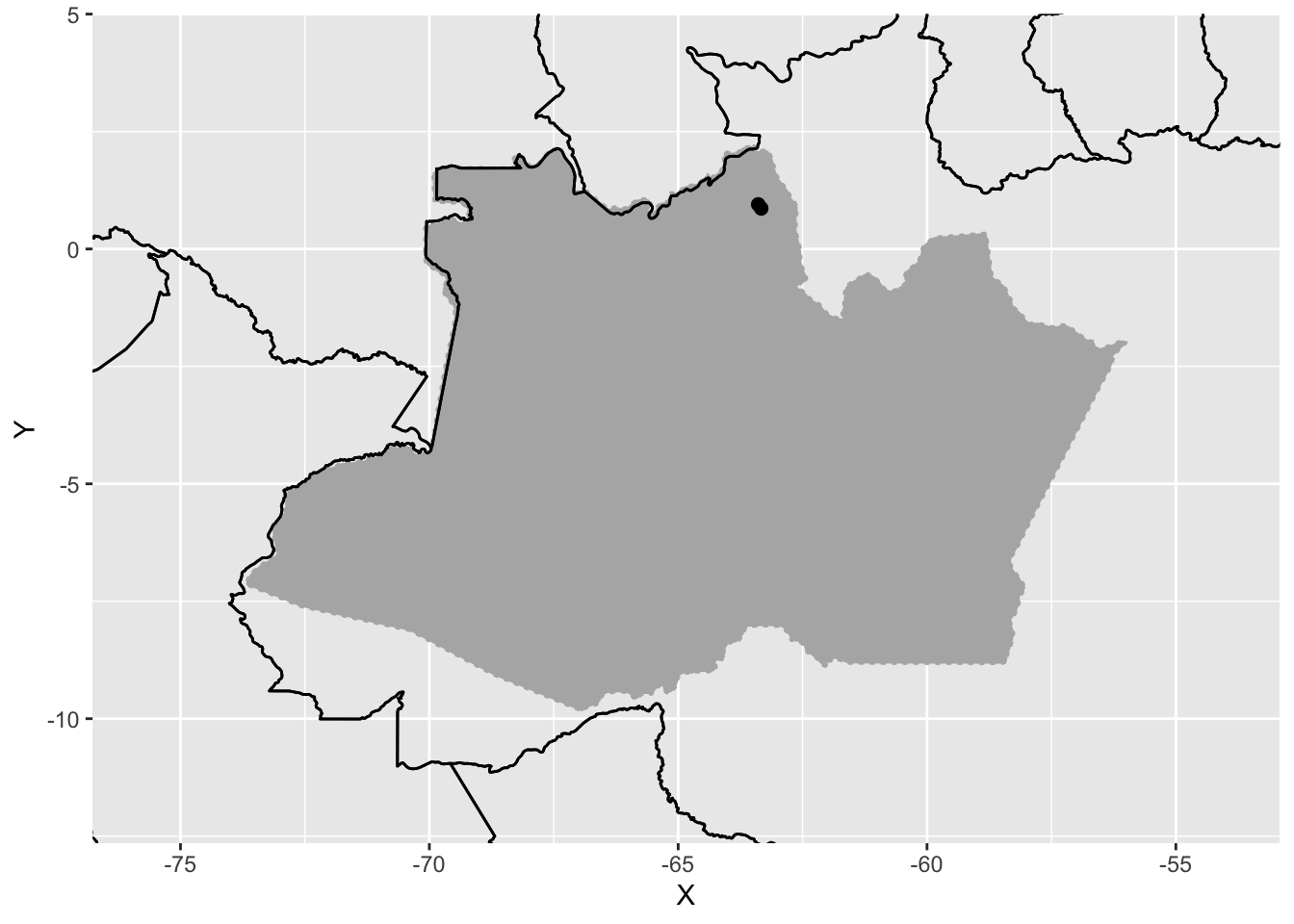
overviewmap <- overviewmap3 +
theme(
plot.background =
element_rect(
fill = "white", linetype = 1,
size = 0.3, colour = "black"
),
axis.line = element_blank(),
axis.text.x = element_blank(),
axis.text.y = element_blank(),
axis.ticks = element_blank(),
axis.title.x = element_blank(),
axis.title.y = element_blank(),
legend.position = "none"
) +
annotate(
x = -63, y = -5, label = "BRAZIL\nAmazonas",
color = "black", geom = "text",
size = 3, fontface = "bold"
)
overviewmap
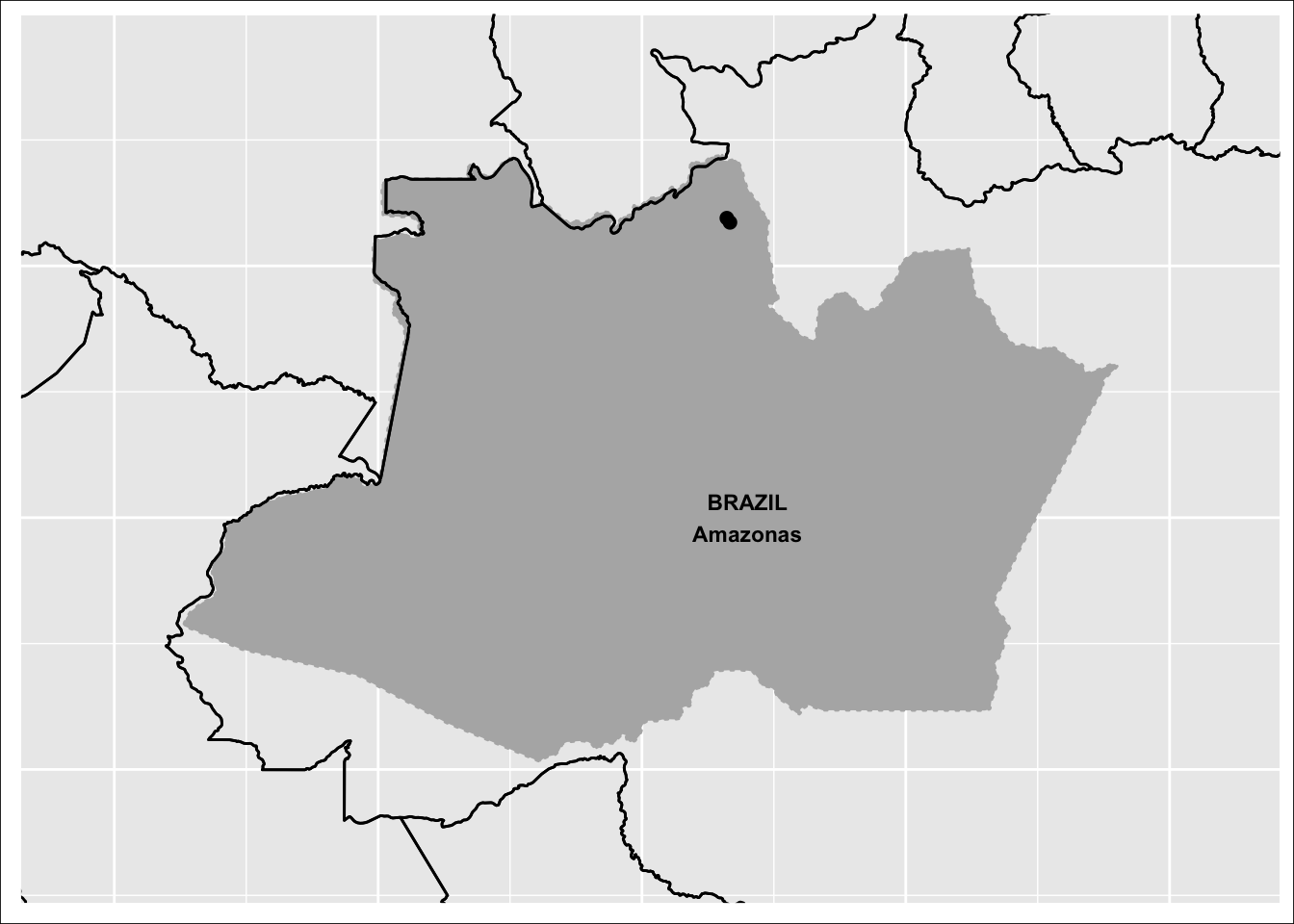
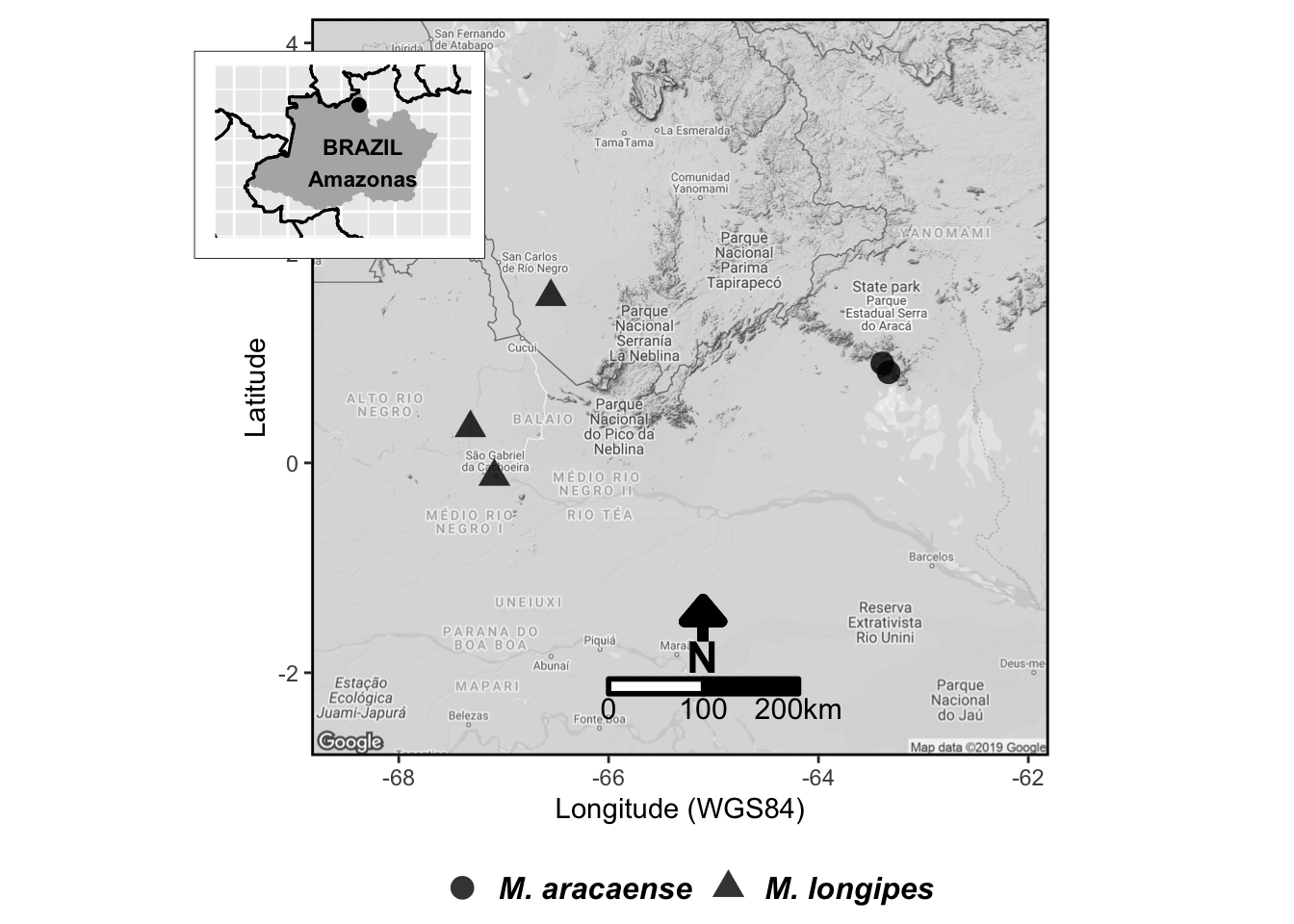
07 Combinam-se os mapas com o pacote R cowplot
final_map <- ggdraw() +
draw_plot(araca_map, 0, 0, 1, 1) +
draw_plot(overviewmap, 0.15, 0.72, 0.225, 0.225)
final_map
08 Salva o mapa final localmente
cowplot::ggsave2(
plot = final_map,
filename = "final_map_Macrolobium_aracaense_ggmap.pdf", dpi = 600, width = 6, height = 6, units = "in"
)
Compare agora a imagem produzida aqui (veja abaixo!) com a publicada em Farroñay et al. (2018).

Referências
Bache, Stefan Milton, e Hadley Wickham. 2020. magrittr: A Forward-Pipe Operator for R. https://CRAN.R-project.org/package=magrittr.
Birk, Matthew A. 2019. measurements: Tools for Units of Measurement. https://CRAN.R-project.org/package=measurements.
Bivand, Roger, Tim Keitt, e Barry Rowlingson. 2021. rgdal: Bindings for the Geospatial Data Abstraction Library. https://CRAN.R-project.org/package=rgdal.
Brunsdon, Chris, e Hongyan Chen. 2014. GISTools: Some further GIS capabilities for R. https://CRAN.R-project.org/package=GISTools.
Farroñay, Francisco, Marisabel U. Adrianzén, Ricardo Oliveira Perdiz, e Alberto Vicentini. 2018. “A new species of Macrolobium (Fabaceae, Detarioideae) endemic on a Tepui of the Guyana shield in Brazil”. Phytotaxa 361 (1): 97–105. https://doi.org/10.11646/phytotaxa.361.1.8.
Henry, Lionel, e Hadley Wickham. 2020. purrr: Functional Programming Tools. https://CRAN.R-project.org/package=purrr.
Pebesma, Edzer. 2021. sf: Simple Features for R. https://CRAN.R-project.org/package=sf.
Robinson, David, Alex Hayes, e Simon Couch. 2022. broom: Convert Statistical Objects into Tidy Tibbles. https://CRAN.R-project.org/package=broom.
Santos Baquero, Oswaldo. 2019. ggsn: North Symbols and Scale Bars for Maps Created with ggplot2 or ggmap. https://github.com/oswaldosantos/ggsn.
Wickham, Hadley. 2019. stringr: Simple, Consistent Wrappers for Common String Operations. https://CRAN.R-project.org/package=stringr.
Wickham, Hadley, Romain François, Lionel Henry, e Kirill Müller. 2021. dplyr: A Grammar of Data Manipulation. https://CRAN.R-project.org/package=dplyr.
Wickham, Hadley, Jim Hester, e Jennifer Bryan. 2021. readr: Read Rectangular Text Data. https://CRAN.R-project.org/package=readr.
Wilke, Claus O. 2020. cowplot: Streamlined Plot Theme and Plot Annotations for ggplot2. https://wilkelab.org/cowplot/.
Xie, Yihui. 2021. knitr: A General-Purpose Package for Dynamic Report Generation in R. https://yihui.org/knitr/.
Xie, Yihui, J. J. Allaire, e Garrett Grolemund. 2018. R Markdown: The Definitive Guide. Boca Raton, Florida: Chapman; Hall/CRC. https://bookdown.org/yihui/rmarkdown.
Xie, Yihui, Christophe Dervieux, e Alison Presmanes Hill. 2021. blogdown: Create Blogs and Websites with R Markdown. https://CRAN.R-project.org/package=blogdown.
- Posted on:
- September 1, 2019
- Length:
- 19 minute read, 3902 words